Frequently Asked Questions
Find answers to top questions about Patentability Search, novelty evaluation, and the patent filing process.
Which databases (NCBI, GenomeQuest, etc.) do you use for sequence searching? ▼
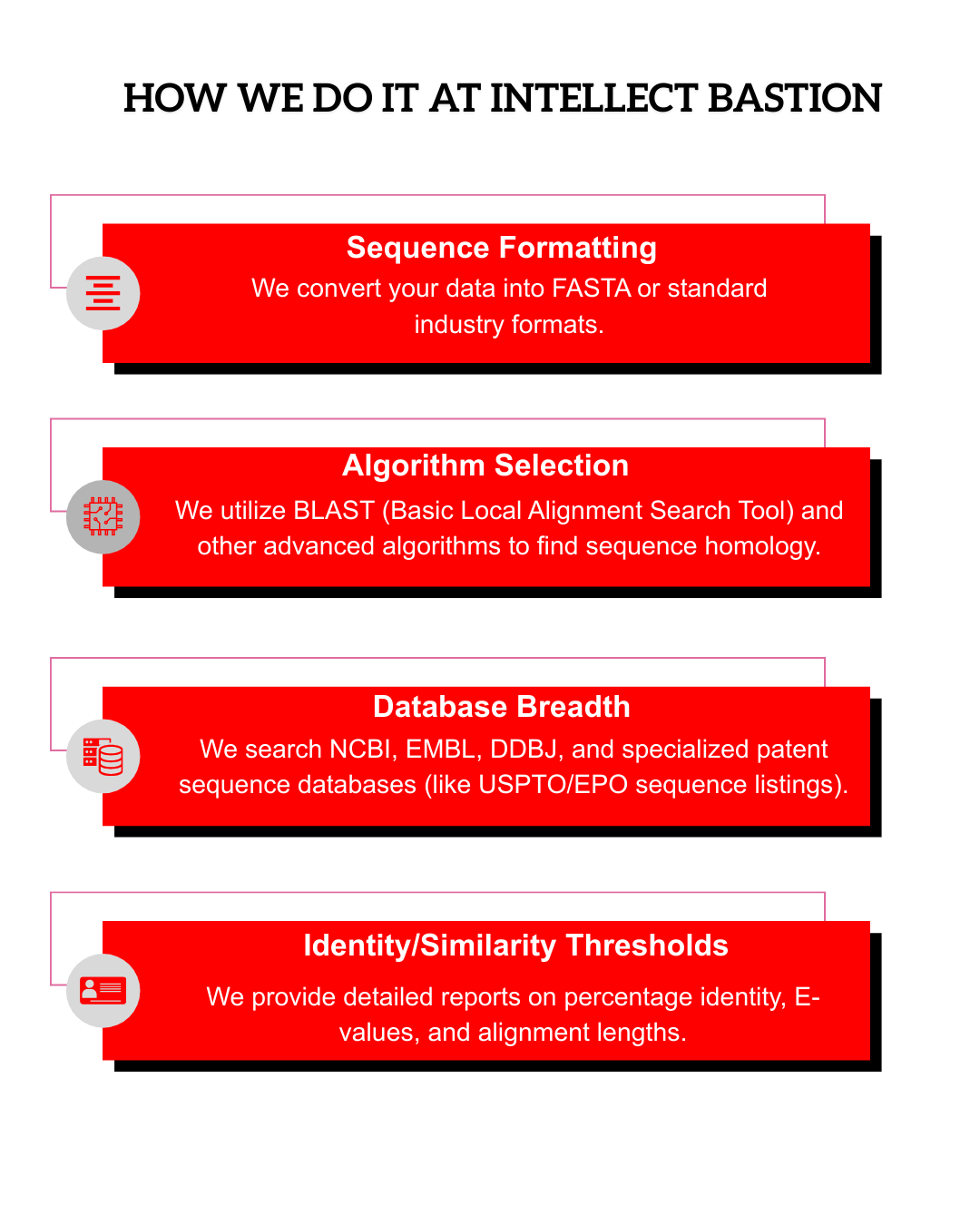
We utilize NCBI (GenBank), EBI, and GenomeQuest, ensuring access to both public genomic data and the "Sequence Listings" found in private patent files.
Can you search for sequences with a certain percentage of "homology" or identity? ▼
Yes. We use BLAST and Smith-Waterman algorithms to find sequences that are 70%, 80%, or 90% identical to your target sequence.
How do you handle searches for modified or synthetic nucleotides? ▼
We can search for non-natural amino acids or modified DNA/RNA bases that are common in modern mRNA and gene-therapy research.
Can you search for "fragments" of a larger gene sequence? ▼
Yes. We perform "Sub-sequence" searches to see if your specific primer, probe, or short peptide is buried within a larger patented gene.
Do you cover sequences found in "Non-Patent Literature" (scientific journals)? ▼
Absolutely. Many genetic breakthroughs are published in journals like Nature or Science months or years before a patent application is made public.
How do you handle search queries for CDRs (Complementarity-Determining Regions) in antibodies? ▼
Yes. We perform targeted searches on the specific hypervariable loops of antibodies, which are the most common areas for patent claims in biologics.
Can you identify patents related to CRISPR or specific gene-editing techniques? ▼
Yes. We can track patents related to specific "Guide RNA" (gRNA) sequences and the Cas9/Cas12 enzyme variations used in gene editing.
How are the results presented (e.g., sequence alignment maps)? ▼
We provide "Alignment Maps" that show your sequence on top of the found patent sequence, highlighting exactly where the "A-T-C-G" or amino acid codes match.
Can you search for "motifs" or specific biological patterns? ▼
Yes. We search for functional patterns (motifs) that might be patented even if the rest of the sequence is entirely different.
How do you account for variations in sequence nomenclature? ▼
Our biological experts account for synonymous codons and the various ways researchers might annotate the same genetic data.